Understanding regulatory network controlling stem cell differentiation
The primary purpose of modeling gene regulatory network for developmental process is to reveal pathways governing the cellular differentiation to specific phenotypes. Knowledge of differentiation network will enable generation of desired cell fates by careful alteration of the governing network by adequate manipulation of the cellular environment. We are developing methods to reconstruct the underlying regulatory architecture of a differentiating cell population from discrete temporal gene and protein expression data. Our primary emphasis is on identifying the governing logic behind network architecture design. The system under investigation is a population of embryonic stem cells differentiating to the pancreatic lineage. The differentiation process is considered to be occurring in a sequence of cascades, enabling modular treatment of the overall ES cell differentiation network. Current work concentrates on a specific stage of pancreatic differentiation, marked by Pdx-1 expression.
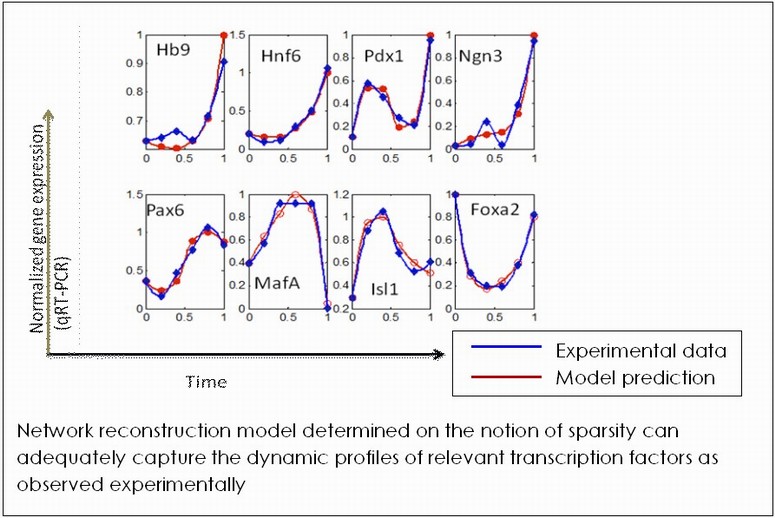
Inducing Embryonic Stem Cells towards Pancreatic Lineage
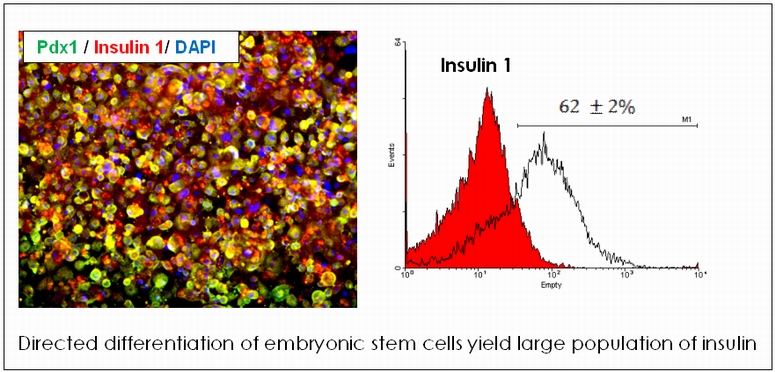
The promise of cellular therapy for type 1 diabetes has inspired the search for transplantable cell sources having the potential to replace the ailing islet cells. Embryonic stem cells (ESC) have emerged as a strong candidate, and generation of large quantities of insulin-producing b cells from these self-renewing ES cells hold tremendous potential for the treatment of diabetes. We are developing a protocol to generate homogenous population of Insulin producing cells triggered by paracrine signals from endothelial cells. In the final stage of maturation, co-culture of the ESC derived pancreatic progenitor cells with endothelial cells is found to have dramatic effect in b cell commitment and insulin 1 expression. More than 60 % of the cell population differentiate into insulin and C-Peptide positive cells, and result in high levels of insulin secretion. We are currently working of analyzing the co-culture system to identify the mechanism by which endothelial cells influence the differentiation.
Adaptive Reduction of Complex Kinetic Mechanisms
Detailed simulation of reactive flow systems using complex kinetic mechanisms, often consisting of hundreds of species and thousands of reactions is a computationally prohibitive task. Hence it is imperative to develop reduced representation of the kinetic models without significant loss of accuracy. We have development an efficient adaptive framework for determination of such reduced models from detailed kinetic networks. The developed framework stems from the understanding that not all reactions are important under all conditions, hence the importance of a reaction or the species involved in the reaction is a strong function of local conditions. Based on this realization we have developed an adaptive reduction framework, where we attempt to identify the set of reactions governing a specific region in the composition space. By spanning the entire composition space we have identified a set of reduced order reaction network models which are governing a defined region in the composition space. The ultimate objective is to integrate the adaptive reduction framework with a detailed flow simulation to enable accurate solution in realistic timeframe